A new program makes 3D protein structure prediction possible; Nobel prize for Chemistry 2024
Proteins are essential for life. For example, to perceive the world beyond ourselves, eyesight is crucial. Inside our eyes, a protein called rhodopsin catches light. Light signals then reach our brain through our neurons. Within each neuron, several proteins work together to transfer light signals to our brain. Like light, medicine also needs proteins to reach their targets in cells before reaching the final target. The target themselves are, more often than not, other proteins located on cell membranes called receptors.
Currently, cancer is the cause of death for one out of three humans. Oncogenesis is associated with genetic mutations. To overcome cancer, we need to understand the nature of “cancer gene products” which are protein molecules produced by mutated genes and which contribute to the development of cancer. These are created from causative genes known as oncogenes in our genome. Originally, oncogene (proto-oncogene) products (proteins) support normally regulated cell growth. But after a mutation in the gene, the protein changes their structure at the atomic level becoming oncogenic, leading to uncontrolled cell-division. Therefore, medical doctors suggest strongly to us not to intake or touch any mutagenic materials around us including cigarettes.
One of major steps towards answering these questions is to understand the 3D structures of proteins. Because such 3D structures provide a cavity or a binding site for a substrate of catalysis by an oncogenic protein, and/or a binding site of a drug. 3D structures are formed by the amino acid sequence comprising the protein. The amino acid sequence is defined by the nucleotide sequence in the genes of our genome. Now we know that we have around 20,000 genes in our genome and that we can predict the amino acid sequence of each protein based on the nucleotide sequence. Until recently, it was impossible to predict the 3D structure of a protein from the amino acid sequence.
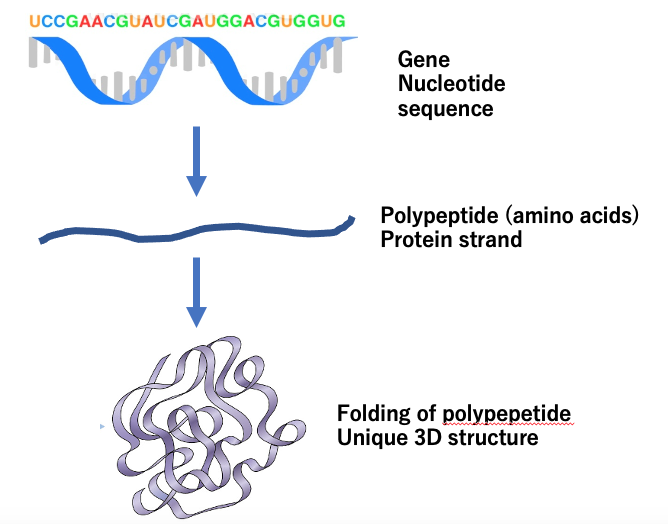
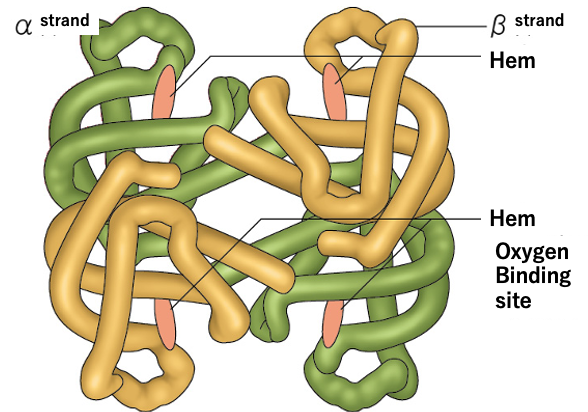
Fig. Upper; an image of protein folding from polypeptide. Lower: An example of 3D structure of protein (hemoglobin).
In 2015, Dr. Baker and Dr. Jumper in a team at Google for Artificial Intelligence achieved partial success in this mission by utilizing deep learning computer systems.
Why was it so difficult for so long to predict 3D structures based on amino acid sequences? A protein is synthesized by a mechanism known as translation within our cells based on messenger RNA after its transcription from DNA. Newly synthesized proteins look like strings of amino acids. These strings then fold into 3D structures as remotely located amino acids interact along the string. For example, a negatively charged amino acid interacts with another positively charged amino acid. Some amino acids, like cysteine, with an SH group in the structure, may interact with another cysteine. The reaction results in the release of H, leading to a bond formed by the so-called S-S bridge. Such complex interactions were once difficult to predict, making it virtually impossible to construct precisely defined 3D structures.
But new AI based techniques make the prediction of these interactions possible. The technique is based on big data of 3D structures formed by about 70,000 proteins obtained with conventional X-ray analysis of protein crystals. The determination of 3D structures through the X-ray analysis of proteins used to take a long time to accomplish, (roughly 10 years for each protein just to make the crystal before analysis). However, the advent of big data have made these predictions much faster and reliable.
The program for this prediction created by Dr. Baker and Dr. Jumper is now named alfa fold 2. It first appeared in a contest for 3D structure prediction named CASP (Critical Assessment of protein Structure Prediction) in 2018. They were able to predict the 3D structure with 90 % accuracy and won the first prize in CASP. Then, in 2024, they were awarded the Nobel prize for Chemistry.
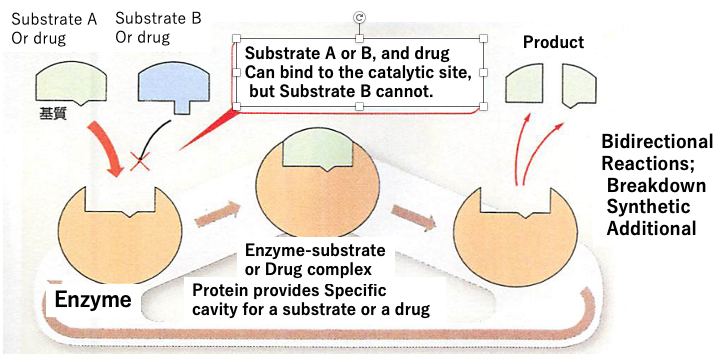
Fig. An enzyme provides a specific binding cavity for substrate A (or drug, compatible forthe binding site) but not for substrate B (incompatible for the binding site).
Since then, the program has become available to the public. Why is the information of 3D structures so valuable? As described in a previous example, drugs bind to target receptors leading to stop the oncogenic reaction by the mutated oncogenic protein. The binding site of the drug is strictly defined by the target protein’s 3D structure at the molecular level. Being able to determine the binding site within the 3D structure allows us to design new drugs tailored to fit the structure leading to stop the oncogenesis. Further we might be able to design a novel protein which lost the oncogenic ability based on the oncogenic gene by using genetic engineering. Now we have a technic to create and introduce this novel protein gene into our body to stop the oncogenesis. In this sense, the alpha fold 2 is an innovation of great significance, leading to the creation of new artificial proteins and opening a future world of new medicine.
Reference; Yusuke Sato, How to use alpha fold 2 (in Japanese), School of Engineering, Tottori Univ. 2018.
